OpenVaccine-COVID-19-mRNA-Vaccine-Degradation-Prediction
Problem statement
The goal of this competition was to predict, for each of the bases (A, C, G or U) in the provided RNA sequences, the reactivity and degradation under certain circumstances such as incubation with or without magnesium in high temperatures or high pH. In summary, we have to predict three continuous values (i.e. regression) for each of the bases in the sequence (i.e. sequence-to-sequence).
The training set consists of 2400 sequences of length 107, the public test set contains 629 sequences of length 107 and the private test set consists of 3005 sequences of length 130. There is thus a difference in the sequence length in the private test data. It is therefore of great importance that the model has built-in invariance to the sequence length.
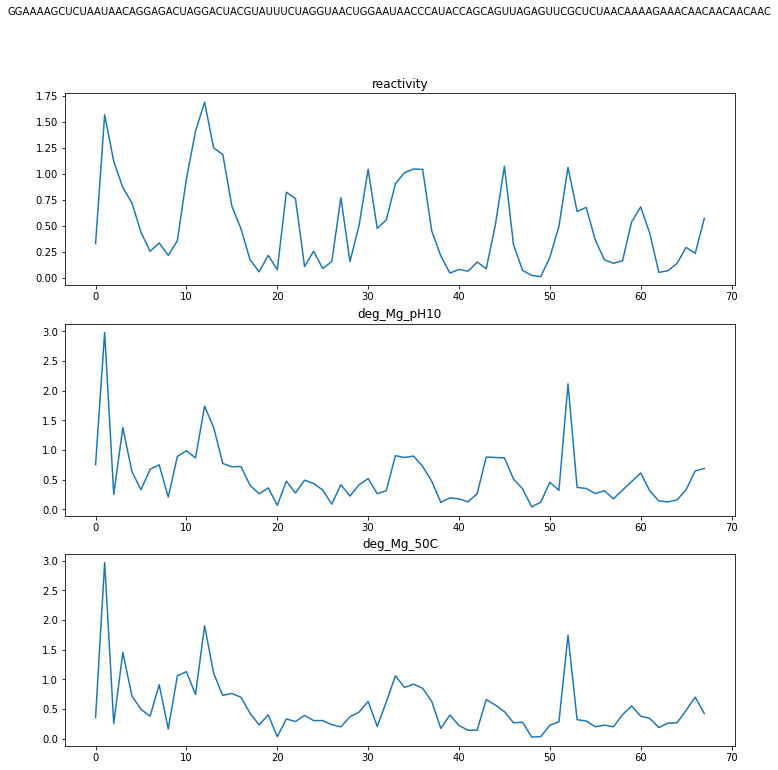
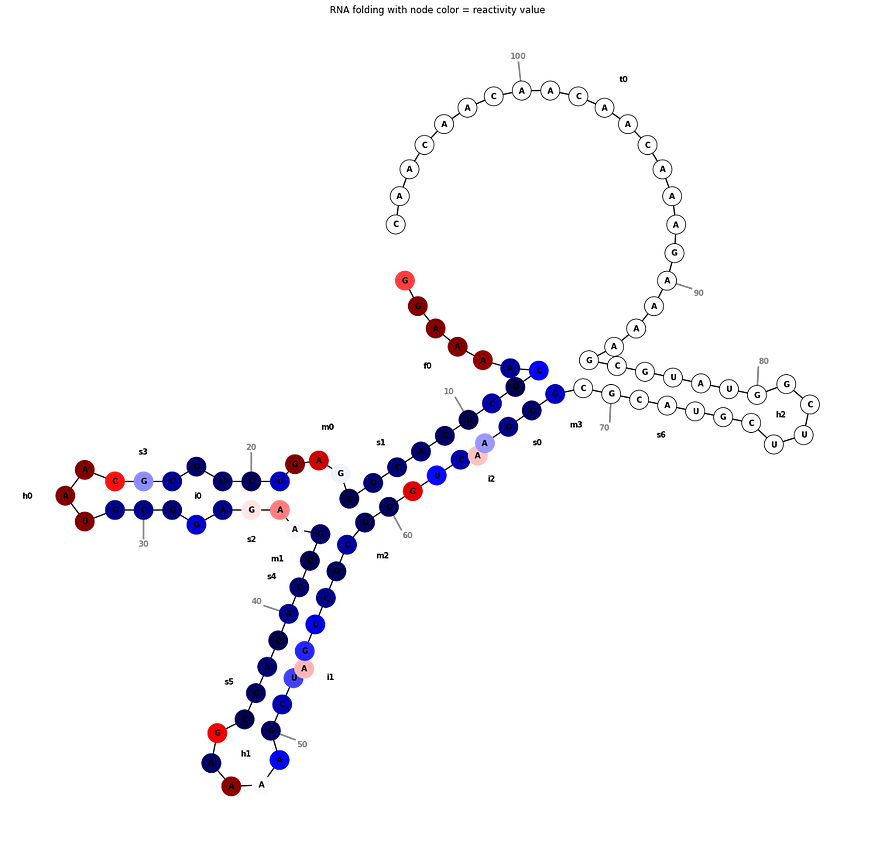
We can also display these target values on the RNA sequence graph directly as done for the reactivity in the example below:
Solution
By looking at the targets, it is clear that a certain value is correlated with its neighbouring values in the time series. Recurrent Neural Networks are a perfect fit for these kinds of situations, as their built-in memory takes into account the nearby predicted values. We learn a representation for each of the bases in our sequence and pass these through 2 (bi-directional) RNN layers. The representation for each node is learned by using both information of the node itself and relationship or edge information. As can be seen in the introduction, the RNA sequence can be represented by a graph, and relations between certain bases in the sequence are present. We use Graph Layers to aggregate this relationship information. Our final solution consists of a blend of 4 different models, in which we vary the RNN layers (LSTM+LSTM, LSTM+GRU, GRU+LSTM and GRU+GRU). Some details are left out from this high-level overview. One important detail is that we pre-trained the network until the RNN layers as an Auto-Encoder. This allowed us to train on the unlabeled test data as well.
Models
- Pytorch RNN
- Keras One-Hot RNN
- Tree models based on NN extracted features
- DeepGNN
- AE pretrained Transformer
- AE pretrained GraphTransformer